Scientists have identified a network of genes that could play an important role in the development of metabolic diseases such as diabetes. The German Mouse Clinic, collaboration partners within the Helmholtz Center Munich and the International Mouse Phenotyping Consortium (IMPC) led the work that is published in 'Nature Communications'.
The development of metabolic diseases like diabetes is a complex process. As well as lifestyle and environmental factors, many different genes are responsible for the pathogenesis of both type 1 and type 2 diabetes. These genes encode information on how to assemble individual proteins that function in glucose metabolism. Many genes that play an important role in the development of human diseases are still unknown.
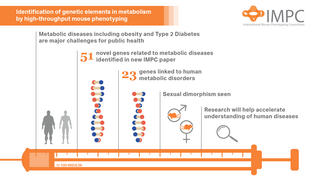
The researchers generated and analyzed metabolic phenotypic data of 2,016 knockout mouse strains under the aegis of the International Mouse Phenotyping Consortium (IMPC). They assessed seven metabolic parameters with diagnostic relevance in human clinical research: fasting basal blood glucose level (T0), area under the curve of blood glucose level after intraperitoneal glucose administration (AUC), plasma triglyceride levels (TG), body mass (BM), metabolic rate (MR), oxygen consumption rate (VO2), and respiratory exchange ratio (RER),which is a measure of whole-body metabolic fuel utilization. This approach uncovered 974 gene knockouts with strong metabolic phenotypes in one or several parameters. 429 of those had no previous link to metabolism and 51 genes remain functionally completely unannotated. The functions of 51 of the discovered metabolic genes in the mouse were hitherto completely unknown. When compared with genome data collected in humans, they found that 23 genes appear to play a role in human diabetes. One of these genes is C4orf22, which appears to be involved in insulin action in participants of the diabetes study "Tübingen Family Study (TÜF)". This needs to be shown for the 51 new genes in the near future.
These candidate genes were also similar in their structure: many had common regulatory elements in promoters. As each regulatory element is composed of several transcription-factor-binding sites, the data reveal an extensive metabolic phenotype-associated network of co-regulated genes. In the future, these new regulatory structures will be investigated and further be explored, to what extent they allow the prediction of gene functions of so-far unknown genes.
Rozman J, Rathkolb B, Oestereicher MA, Schütt C, Ravindranath AC, Leuchtenberger S, Sharma S, Kistler M, Willershäuser M, Brommage R, Meehan TF, Mason J, Haselimashhadi H; IMPC Consortium, Hough T, Mallon AM, Wells S, Santos L, Lelliott CJ, White JK, Sorg T, Champy MF, Bower LR, Reynolds CL, Flenniken AM, Murray SA, Nutter LMJ, Svenson KL, West D, Tocchini-Valentini GP, Beaudet AL, Bosch F, Braun RB, Dobbie MS, Gao X, Herault Y, Moshiri A, Moore BA, Kent Lloyd KC, McKerlie C, Masuya H, Tanaka N, Flicek P, Parkinson HE, Sedlacek R, Seong JK, Wang CL, Moore M, Brown SD, Tschöp MH, Wurst W, Klingenspor M, Wolf E, Beckers J, Machicao F, Peter A, Staiger H, Häring HU, Grallert H, Campillos M, Maier H, Fuchs H, Gailus-Durner V, Werner T, Hrabe de Angelis M. (2018): Identification of genetic elements in metabolism by high-throughput mouse phenotyping Nat Commun. 2018